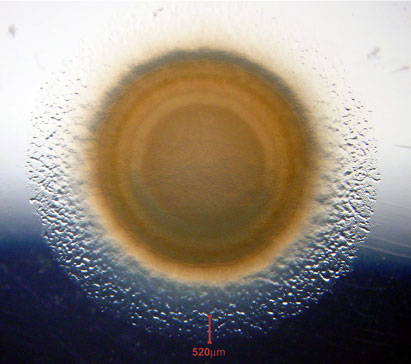
elaborates a swarm whose cells are constantly moving. The movements are well organized
and the cells never seem to jam up even though collisions are frequent. They move constantly
in order that all cells have access to oxygen from the atmosphere above and to carbon,
nitrogen, and phosphate nutrients from the agar medium below. The long, rod-shaped
cells power movement only in the direction of their long axis. Cells move this way for
hydrodynamic reasons and because their motility engines are located at the cell poles.
For S-motility there are type IV pili at the front to pull cells forward. For A-motility there are
slime secretion engines at the back to push cells forward. Forward movement is
punctuated with reversal and equal movement in the opposite direction. An oscillator,
known as the "pacemaker", causes each cell to reverse direction every 8 – 9 min (Kaiser
& Warrick, 2011). The periodic reversals are essential for M. xanthus to swarm; if
reversals are blocked, a swarm cannot expand (Wu et al., 2009). Cell flexibility
(Wolgemuth, 2005, Bui et al., 2009) and frequent reversals prevent collisions between
cells from becoming traffic jams. Swarm cells appear to move at random without
attenuating the movement of their neighbors. The product is a circular swarm (see photo)
that expands at a constant rate, unlike a colony that expands at an ever decreasing rate as
cells compete for nutrient and oxygen. Even when swarming at rather high density, an 0.5
mm wide ring of cells at the swarm edge are growing as fast as cells can grow in aerated
liquid culture. The organized behavior of an M. xanthus swarm in its steady state supports
rapid growth.
Swarming
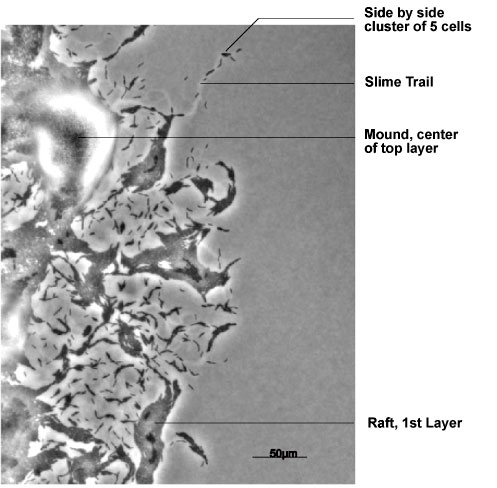
Swarms build 2 sorts of well-organized, multicellular structures at the growing
and expanding edge of the swarm: rafts and mounds. Both structures are identified in the
figure. Rafts are dense, single-layered, rectangular assemblies of hundreds of cells that
have their long axes in parallel with each other. Although raft cells are packed together,
individual cells are able to slide smoothly along side their neighbors, and to reverse their
gliding direction independently of each other. Mounds are round, multi-layered
assemblies of thousands of cells. The layers are nested one on top of another. Due to their
layering, mound cells move in 2 different ways. They can move rapidly from one layer to
the next, either up or down. Alternatively, mound cells can move more slowly to a new
position within the same layer. Mounds are observed to be built from the bottom up, and
for that reason cells in the top layer will have been mound residents longer than any other
cells (Kaiser & Warrick, in preparation).
Each cell has its own pacemaker, an oscillator that causes it to reverse its gliding
direction regularly, every 8-9 min (Kaiser & Warrick, 2011). Recently evidence for a
signal has come to light which brings the pacemakers in a pair of cells that are signaling
to each other, to the same phase of their reversal cycle. The signaling cells become
synchronized with each other. Moreover, once synchronized, 2 pacemakers remain
synchronized. The signal was discovered when viewing time-lapse movies of a mound.
After a 1 hr delay, all cells in the top (5th) layer of a mound "exploded". All of a sudden
they increased their speed of movement 10-fold to 12.5 μm/min, then within a minute
decreased their speed back to the 1.2 μm/min speed average. The timing signal is thought
to spread by transient aligned contact between pairs of adjacent cells. Accordingly, the
hour's delay is the time required to spread the signal among all cells in the mound's top
layer.
The signal is thought to be transferred from one cell to another via multi-protein
complexes called "focal adhesions" (Mignot et al., 2005, Mignot et al., 2007). Focal
adhesions are found as a series of discrete foci running along one side of a cell from its
leading end to just forward of mid-cell. The 15 or more proteins that characterize an
adhesion seem to be suited for signaling (Kaiser & Warrick, in preparation), because
they have the capacity to bind each other in pairs (Nan et al., 2010). Moreover, they are
localized in each of the membrane-bound compartments of M. xanthus cells. Proceeding
inward, the compartments are defined by the outer membrane, the periplasm, the outer
surface of the peptidoglycan sacculus, the inner membrane, and finally the cytoplasm of
these Gram-negative bacteria. Accordingly, the signal could link oscillating methylated-
FrzCD in the pacemaker of one cell mechanically through pairs of protein 1 transiently
bound to protein 2 links to CglB (Hodgkin & Kaiser, 1979a) – one of the 15 motility proteins
found in an adhesion – on the surface of that cell. It is proposed that CglB on the surface
of the first cell would assemble with CglB on the surface of the second cell. From CglB,
the signal would link through the same series of protein 1• protein 2 pairs until it reached
FrzCD in the pacemaker of the second cell. When completed, the series of signal links
would bring the pacemakers of both cells to the same, average phase of their oscillatory
cycles. We also study the cell-to-cell signals involved in fruiting body development.