by 2 distinct genetic systems known as the adventurous (A) and social (S) motility systems (Hodgkin & Kaiser, 1979a) (Hodgkin & Kaiser, 1979b). S-motility is active between cells that
are separated from each other, while A-motility allows cells to move independently. Mutations
in the S-system genes do not interfere with A-motility, and vice-versa. A single mutation in
the mgl locus, however, interferes with both types of movement in a swarm.
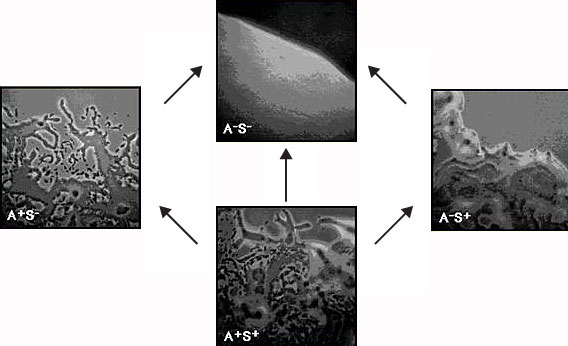
Pictured above are the edges of colonies of M. xanthus grown on agar. The top frame
shows the smooth edge of a colony which has neither A nor S motility, and therefore
does not swarm at all; this is characteristic of strains with a single mutation in the
mgl locus, as well as of strains with one mutation in each of the two motility systems.
The left frame depicts a colony in which only A-motility is active; the right frame
shows a colony in which only S-motility is active. Finally, the bottom frame shows a
wild-type colony in which both A and S-motility are active.
S-MOTILITY
The engines of S-motility, which occupy the leading end of the cell, are polar type IV
pili. Their structure and ability to retract forcefully have been reviewed (Nudelman &
Kaiser, 2004). Table 1 lists the locations and functions of the pilus proteins. In addition
to pili, S-motility requires polysaccharide fibrils described by (Behmlander & Dworkin,
1994). Evidence that the tips of M. xanthus pili bind large fibrils that are enveloping
clusters of cells is summarized in (Kaiser & Warrick, in preparation).
Table 1: S-motility proteins
| Protein (a) |
Function | Cellular Localization | Clustered (b) |
Conserved in multiple species (c) |
| PilA | Pilin, monomer unit of the pilus filament |
Assembled into the pilus fiber Stored in the Inner Membrane |
+ | + |
| PilB | Pilus extension | Inner membrane | + | + |
| PilT | Pilus retraction | Inner membrane | + | + |
| PilC | Unknown | Inner membrane | + | + |
| PilD | PilA leader peptidase |
Inner membrane | + | + |
| PilG | ABC Transporter | Periplasm | + | |
| PilH | ABC Transporter | Periplasm | + | |
| PilI | ABC Transporter | Periplasm | + | |
| PilM | ATPase | Inner membrane | + | + |
| PilN | Unknown | Periplasm | + | + |
| PilO | Unknown | Periplasm | + | + |
| PilP | Unknown | Anchored in outer membrane |
+ | + |
| PilQ | Secretin | Outer membrane | + | + |
| PilR | Regulates transcription of pilA |
+ | - | |
| PilS | Two-component sensor for pilR |
+ | - | |
| PilR1 | Transcriptional regulator |
+ | - | |
| PilS1 | Two-component sensor for pilR1 |
+ | - | |
| Tgl | Secretin assembly factor |
Outer membrane | - | - |
(a) Pil genes named as in P. aeruginosa.
(b) A set of 16 contiguous genes in M. xanthus.
(c) M. xanthus, P. aeruginosa, N. gonorrhoeae, N. meningitigis and
Synechocystis PCC 6803 (Nudleman and Kaiser 2004).
A-MOTILITY & SLIME SECRETION
A-motility is used to separate the 2 daughter cells after they have divided; it is used for
swarming, and for the construction of fruiting bodies. Moreover, it is regulated by the
same reversal switch as the type IV pili (Kaiser, 2008). Many slime trails are evident at
the edge of the swarm, and cells are always found on a trail. Hodgkin isolated mutants
specifically deficient in A-motile gliding, and established that A-motility involved a
different set of genes from the type IV pilus-dependent S-motility. The trail elongates at
exactly the rate at which the cell that moves forward is depositing the trail. The trail
begins at the lagging end of each cell, and is observed to lengthen at the same rate as the
leading end of the cell advances. High resolution transmission electron microscopy (EM)
revealed long, narrow, amorphous filaments, thought to be extruded slime, emerging
from one end of the cell (Wolgemuth et al., 2002). Moreover, by phase contrast visible
light microscopy, filaments were seen to emerge only from one end of each cell (Yu &
Kaiser, 2007). EM also revealed more than 500 thick walled rings, 80% of which are
distributed between the two ends of the cell. The slime filaments emerge in the vicinity of
the polar rings. The rings were seen to be remarkably uniform, having an outer diameter
(OD) of 13 nm and an inner diameter (ID) of 6.5 nm. Rings are interpreted to be end-on
views of secretory nozzles that from the side are amphora-shaped.
| ADVENTUROUS MOTILITY The edge of a colony displaying adventurous motility |
|
| (600x time-lapse, 10 frames/sec) | |
The narrow filaments of slime observed in the EM (Wolgemuth et al., 2002) were
observed to fuse laterally with each other to form the single broad ribbon that is visible
with phase-contrast illumination (Yu & Kaiser, 2007). At any particular moment, both
EM and light microscopy clearly show that the filaments are present at only one of the
two cell poles (Wolgemuth et al., 2002) (Yu & Kaiser, 2007). Unipolarity of the extruded
filaments of slime matches the instantaneous unidirectional movement of cells. The
phase-bright trail that is left behind when wild type cells move is extruded slime left
behind on the agar. The opposite end has no filament even though it has more than a
hundred nozzles (Wolgemuth et al., 2002). Therefore, many of the 400 polar nozzles are
not secreting propulsive slime at any instant that a cell is moving. Approximately 40% of
all nozzles can't secrete because they are situated at the leading end of the cell, the wrong
end for pushing with slime secretion. The next time the cell reverses its gliding direction,
these polar nozzles will secrete slime. The remaining 20% of the walled rings are found
on the side of each cell, and they are secreting slime for the capsule that protects M.
xanthus against lysis from without by its own secreted digestive enzymes. This is the
vital function of the capsule in many Gram-negative bacteria (Whitfield, 2006), and it
explains why A-motility mutant have never been found that fail to secrete slime.
The realization that nozzles are secretion pores that can be turned on and off
coincides with the identification of 2 large classes of A-motility genes (Yu & Kaiser,
2007). Half the knock-out mutants produced by transposon-insertion were secreting slime
from both ends simultaneously, and all these mutants were non-motile. Because the
bipolar secretion mutants lack motility, uni-polar secretion is needed for A-motility.
Indeed, the other half of Yu's transposon-insertion mutants were secreting slime from 1
end only, and they retained A-motility. Although they are motile and they have normal
reversal frequencies, these mutants move less often; they expand their swarms at
the same fractional rate that they moved. For this reason, they were called pgl for partial
gliding motility (Yu & Kaiser, 2007). Most of these pgl insertion mutations were found in
genes that encoded a sugar transferase enzyme or a regulator (Yu & Kaiser, 2007), strongly
suggesting roles in polysaccharide biosynthesis. This suggestion is supported by earlier
observations of (Spormann & Kaiser, 1999). Spormann showed that mutants with
deletion of mglA and/or of mglB reversed their gliding direction 10-times more frequently
than wildtype. Accordingly, they only ever moved a small fraction of a cell length before
reversing again (Spormann & Kaiser, 1999). Moreover, the extra reversals were jerky and
noticeably more abrupt than wildtype. These observations fit the following statistical
expectation for mutants that secrete slime simultaneously from nozzles at both poles.
Since the bipolar mutants are common and since there are several hundred nozzles at both
poles in the bipolar mutants, one would expect by chance and from moment to moment
there would be a few more active nozzles at one end than at the other, because the
reversals are not pacemaker-induced, but chance-induced. Being chance-induced, one
would also expect the end with the more nozzles would switch more often and to switch
more abruptly. Cuthbertson et al. have reviewed the general biochemistry of capsular
slime secretion, including the description of secretion genes in M. xanthus (Cuthbertson
et al., 2009). Considering that the capsule is essential, it seems likely that M. xanthus
augmented its capacity for capsule synthesis to propel its gliding motility by adding more
secretory nozzles to both ends of each cell, and by activating slime secretion one end at a
time to generate polarized secretion and directional gliding movement.